research
Spatial-extent inference
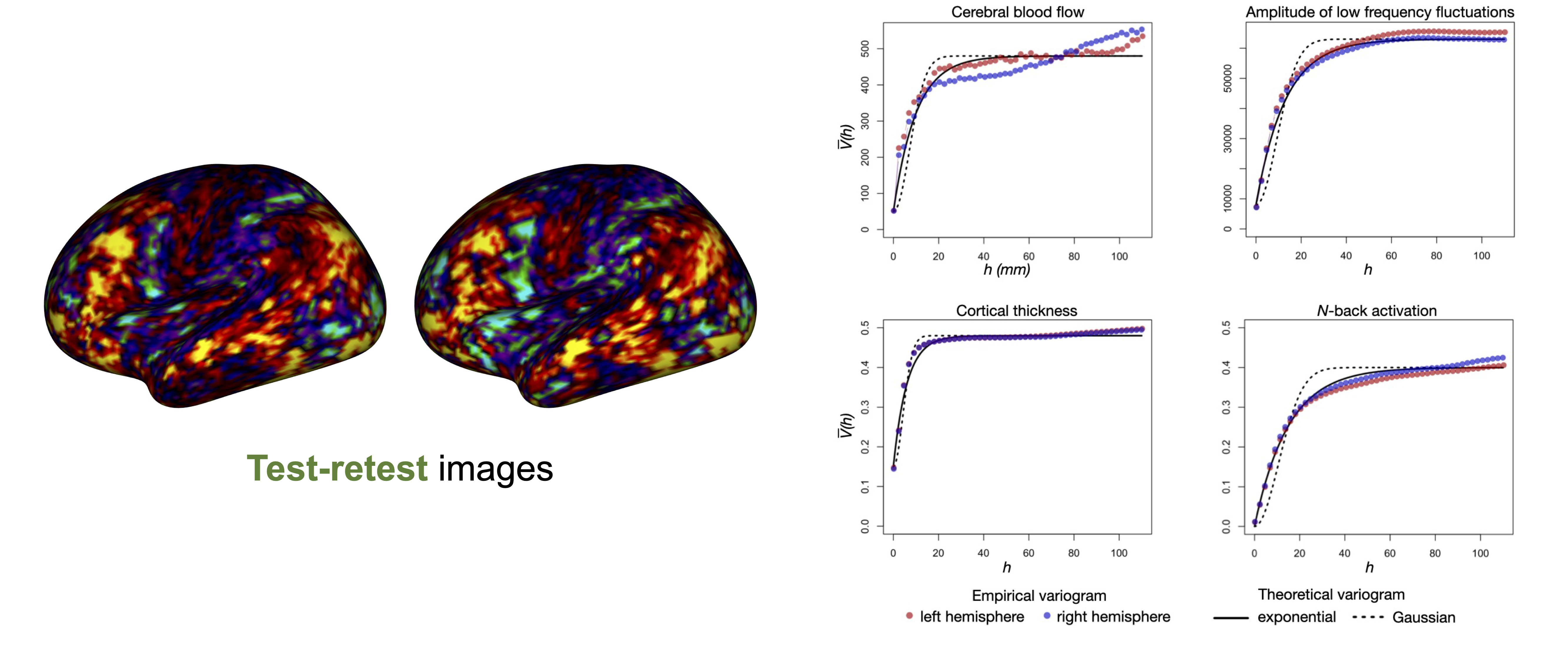
References
-
Pan, R., Weinstein, S. M., Tu, D., Hu, F., Tanriverdi, B, Zhang, R., Baller, E.B., … Park, J.Y (2025). Mapping individual differences in intermodal coupling in neurodevelopment. Imaging Neuroscience.
-
Pan, R., Dickie, E. W., Hawco, C., Reid, N., Voineskos, A. N., Park, J. Y. (2024). Spatial-extent inference for testing variance components in reliability and heritability studies. Imaging Neuroscience.
-
Weinstein, S. M., Vandekar, S. N., Baller, E. B., Tu, D., Adebimpe, A., Tapera, T. M., … Park, J. Y. (2022). Spatially-enhanced clusterwise inference for testing and localizing intermodal correspondence. NeuroImage, 264, 119712.
-
Park, J. Y., Fiecas, M. (2022). CLEAN: Leveraging spatial autocorrelation in neuroimaging data in clusterwise inference. NeuroImage, 255, 119192.
-
Park, J. Y., Fiecas, M. for the Alzheimer’s Disease Neuroimaging Initiative. (2021). Permutation-based inference for spatially localized signals in longitudinal MRI data. NeuroImage, 239, 118312.
Integration of multi-view data
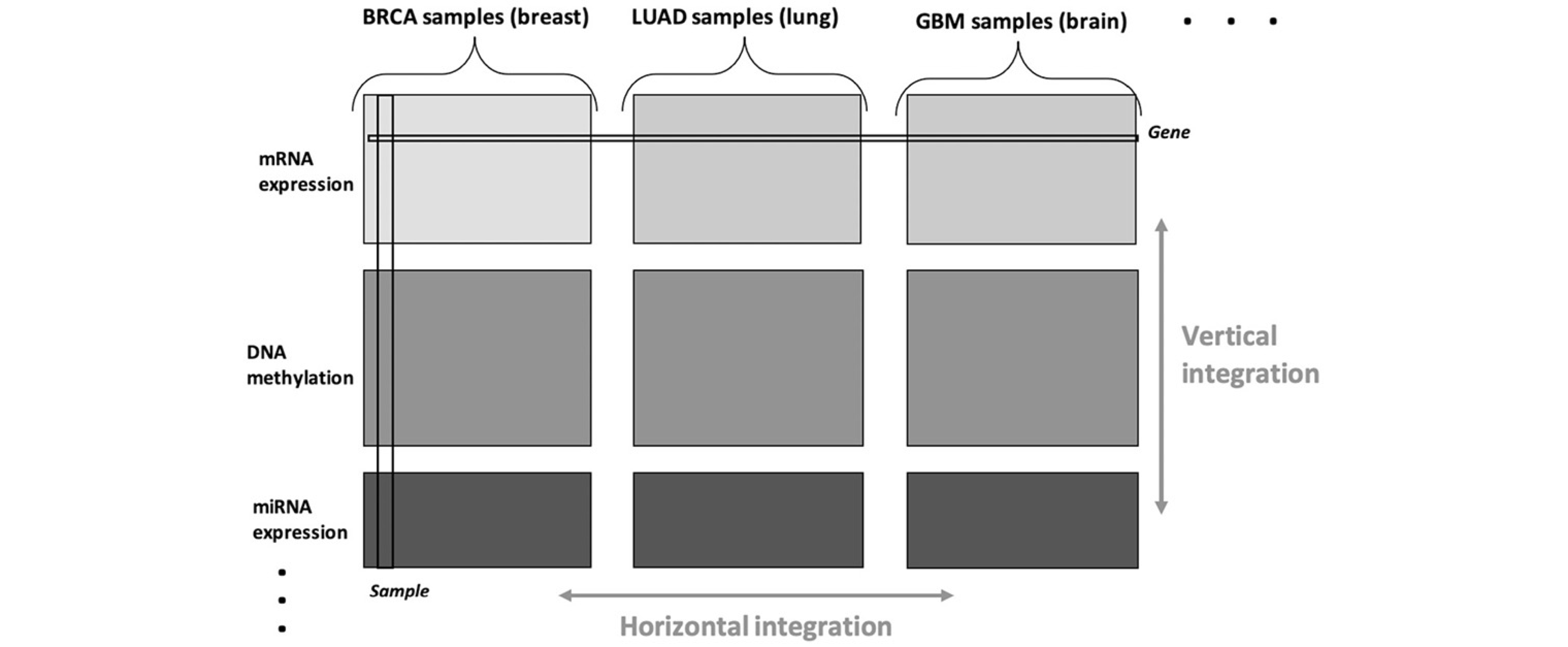
References
-
Lock, E. F., Park, J. Y., Hoadley, K. A. (2022). Bidimensional linked matrix factorization for pan-omics pan-cancer analysis. The Annals of Applied Statistics, 16(1), 193.
-
Park, J. Y., Lock, E. F. (2020). Integrative factorization of bidimensionally linked matrices. Biometrics, 76(1), 61-74.
Data harmonization
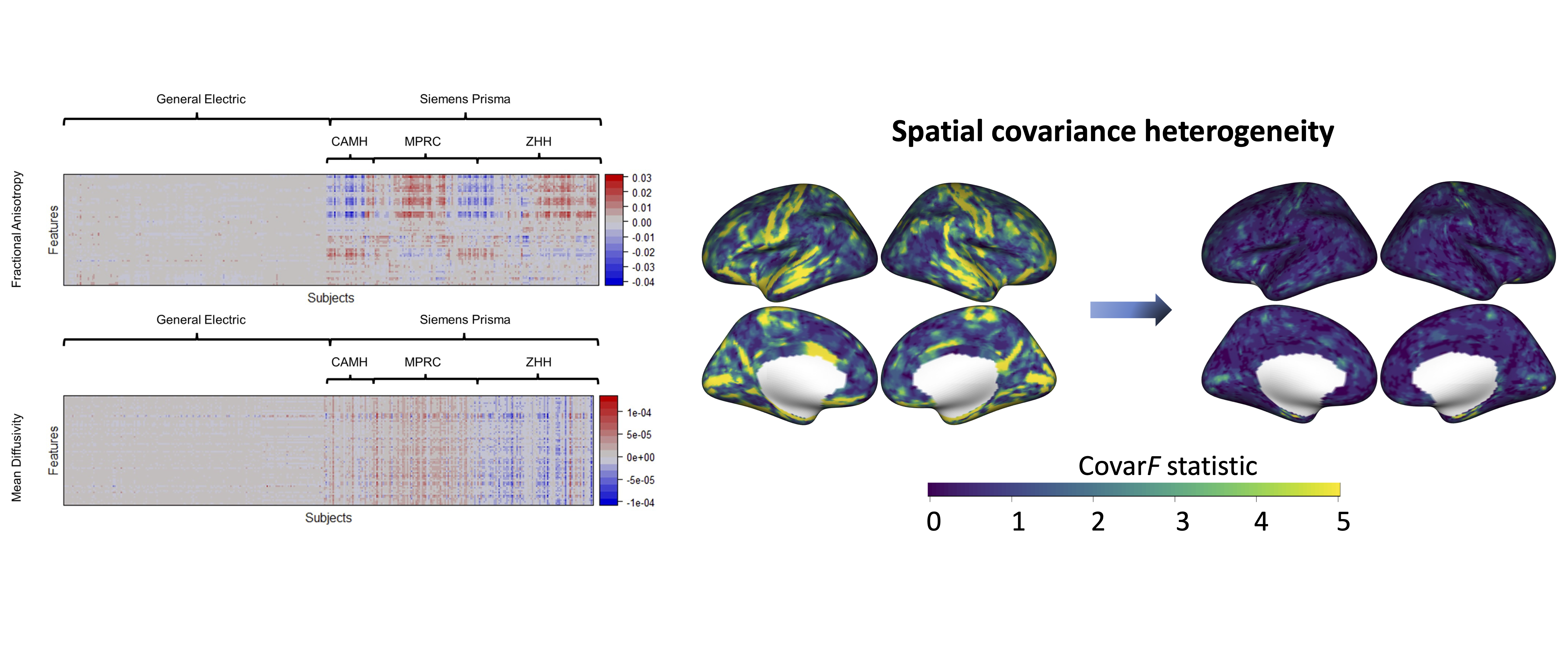
References
-
Zhang, R., Tuzhilina, E., Park, J. Y. (2026). Sparse covariate-driven factorization of high-dimensional brain connectivity with application to site effect correction. Arxiv, Arxiv:2601.09525.
-
Zhang, R., Chen, L., Oliver, L. D., Voineskos, A. N., Park, J. Y. (2024). SAN: mitigating spatial covariance heterogeneity in cortical thickness data collected from multiple scanners or sites. Human Brain Mapping, 45:e26692.
-
Zhang, R., Oliver, L. D., Voineskos, A. N., Park, J. Y. (2023). RELIEF: A structured multivariate approach for removal of latent inter-scanner effects. Imaging Neuroscience, 1, 1-16.
Time series
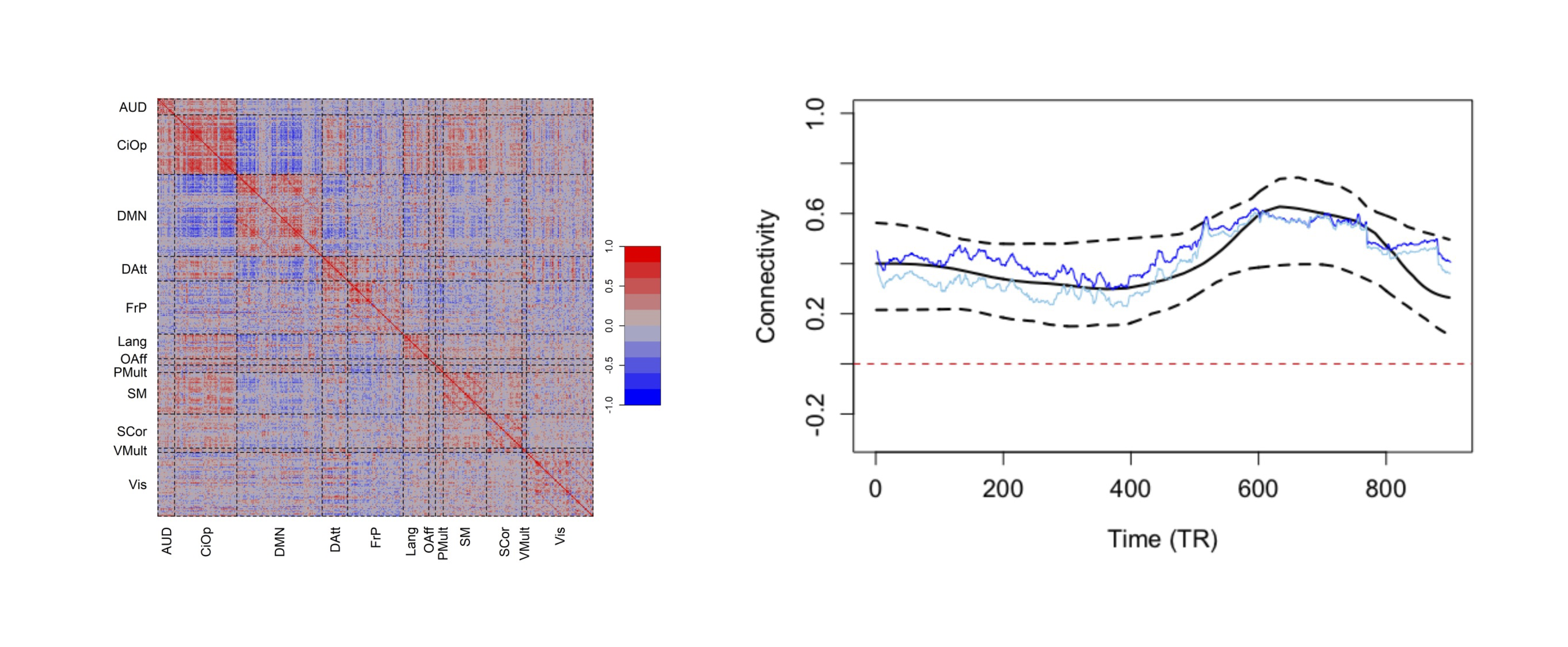
References
-
Veitch, D., He, Y., Park, J. Y. (2023). Rank-adaptive covariance changepoint detection for estimating dynamic functional connectivity from fMRI data. arXiv, arXiv:2309.10284.
-
Park, J. Y., Polzehl, J., Chatterjee, S., Brechmann, A., Fiecas, M. (2020). Semiparametric modeling of time-varying activation and connectivity in task-based fMRI data. Computational Statistics & Data Analysis, 150, 107006.
Statistical genetics
References
-
Tian, Y., Felsky, D., Gronsbell, J., Park, J. Y. (2025). Leveraging multimodal neuroimaging and GWAS for identifying modality-level causal pathways to Alzheimer’s disease. Imaging Neuroscience
-
Park, J. Y., Wu, C., Basu, S., McGue, M., Pan, W. (2018). Adaptive SNP-set association testing in generalized linear mixed models with application to family studies. Behavior genetics, 48, 55-66.
-
Wu, C., Park, J. Y., Guan, W., Pan, W. (2018, September). An adaptive gene-based test for methylation data. In BMC proceedings (Vol. 12, pp. 85-89). BioMed Central.
-
Park, J. Y., Wu, C., Pan, W. (2018). An adaptive gene-level association test for pedigree data. BMC genetics, 19(1), 39-43.